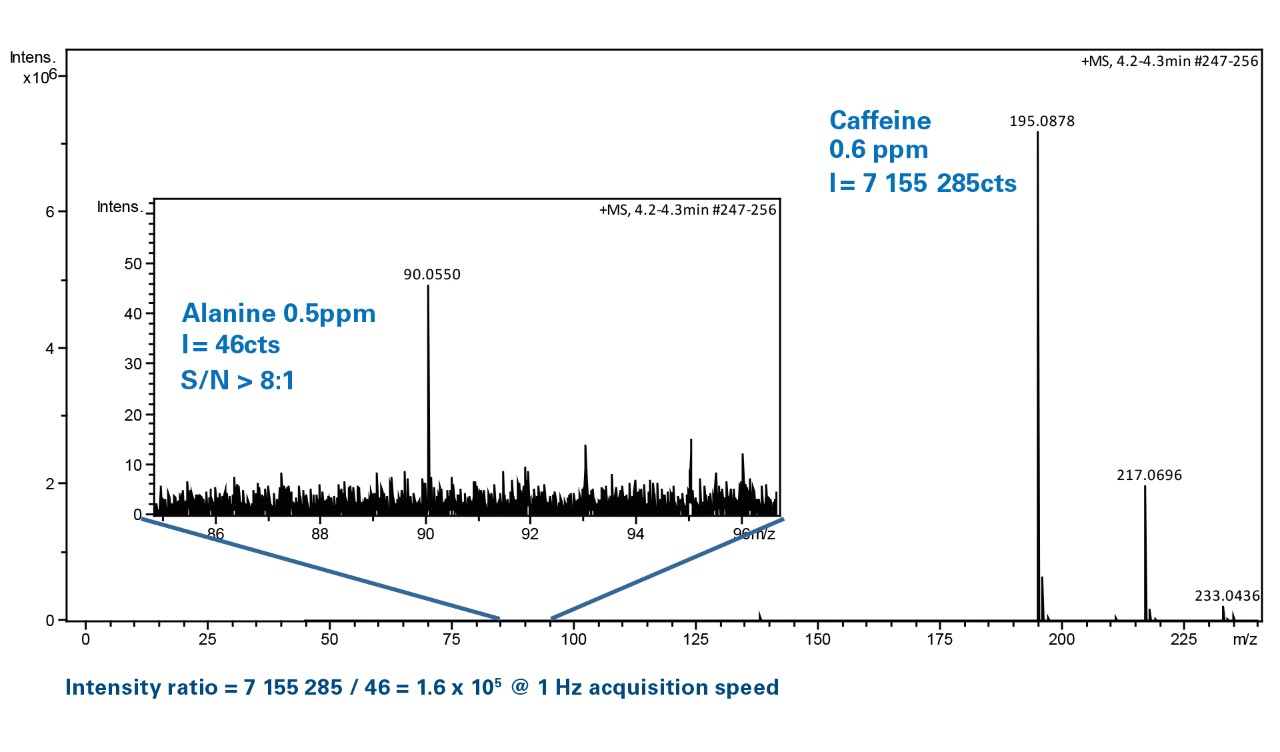
3.3. Estimating the linear range | MOOC: Validation of liquid chromatography mass spectrometry (LC-MS) methods (analytical chemistry) course

Improved Dynamic Range, Resolving Power, and Sensitivity Achievable with FT-ICR Mass Spectrometry at 21 T Reveals the Hidden Complexity of Natural Organic Matter | Analytical Chemistry

Expanding the linear dynamic range for quantitative liquid chromatography-high resolution mass spectrometry utilizing natural isotopologue signals - ScienceDirect

Improved Quantitative Dynamic Range of Time-of-Flight Mass Spectrometry by Simultaneously Waveform-Averaging and Ion-Counting Data Acquisition | Journal of The American Society for Mass Spectrometry
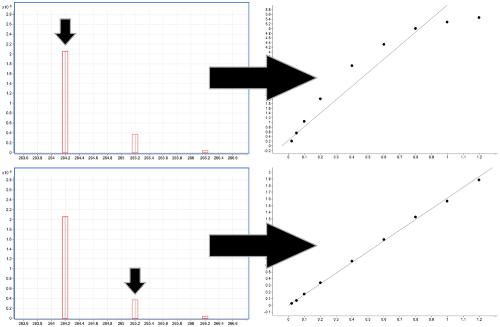
Increasing the linear dynamic range in LC–MS: is it valid to use a less abundant isotopologue?,Drug Testing and Analysis - X-MOL
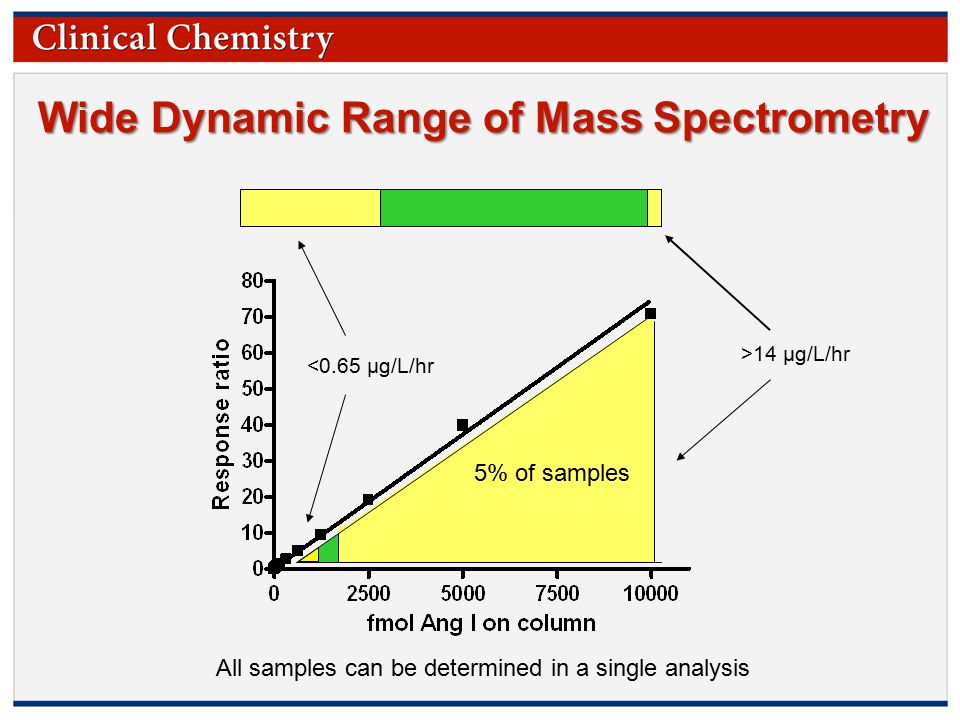
Copyright 2009 by the American Association for Clinical Chemistry Plasma Renin Activity by Liquid Chromatography– Tandem Mass Spectrometry (LC-MS/MS): - ppt download
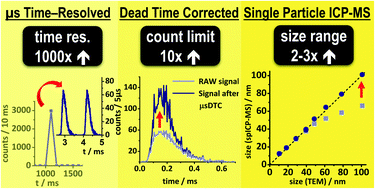
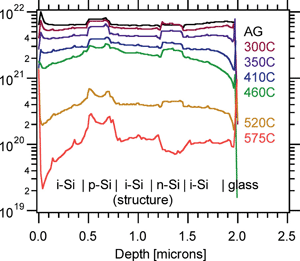
Single particle inductively coupled plasma mass spectrometry: investigating nonlinear response observed in pulse counting mode and extending the linear dynamic range by compensating for dead time related count losses on a microsecond

Extending mass spectrometry access to the full plasma proteome dynamic... | Download Scientific Diagram

Comparison of linear intrascan and interscan dynamic ranges of Orbitrap and ion‐mobility time‐of‐flight mass spectrometers - Kaufmann - 2017 - Rapid Communications in Mass Spectrometry - Wiley Online Library
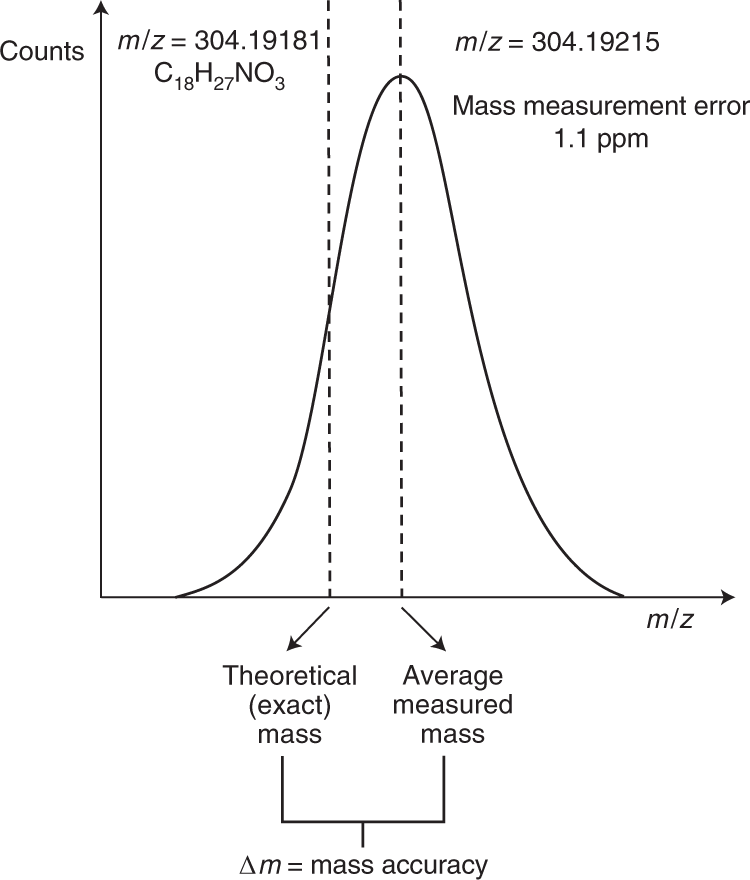
Ultra-high-performance liquid chromatography high-resolution mass spectrometry variants for metabolomics research | Nature Methods

Improving the success rate of proteome analysis by modeling protein-abundance distributions and experimental designs | Nature Biotechnology

Comparison of linear intrascan and interscan dynamic ranges of Orbitrap and ion‐mobility time‐of‐flight mass spectrometers - Kaufmann - 2017 - Rapid Communications in Mass Spectrometry - Wiley Online Library

Figure 2 from Mass Spectrometry-Based Identification of Muscle-Associated and Muscle-Derived Proteomic Biomarkers of Dystrophinopathies. | Semantic Scholar

Figure 5 from A microelectromechanical systems-enabled, miniature triple quadrupole mass spectrometer. | Semantic Scholar

Improved Dynamic Range, Resolving Power, and Sensitivity Achievable with FT-ICR Mass Spectrometry at 21 T Reveals the Hidden Complexity of Natural Organic Matter | Analytical Chemistry
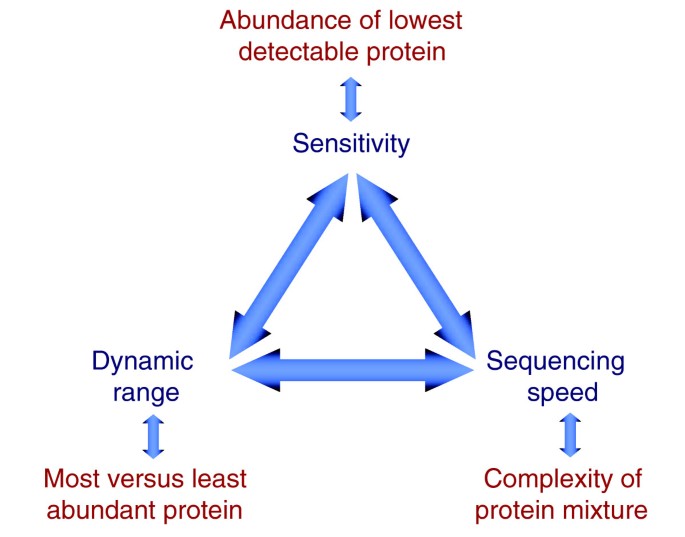
Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system | Genome Biology | Full Text

Figure 1 from Mass Spectrometry-Based Identification of Muscle-Associated and Muscle-Derived Proteomic Biomarkers of Dystrophinopathies. | Semantic Scholar